One of the principal features used in analytic databases is table partitioning. This feature is so frequently used because of its ability to significantly reduce query latency by allowing the execution engine to skip reading data that is not necessary for the query. For example, consider a table of events partitioned on the event time using calendar day granularity. If the table contained 2 years of events and a user wanted to find the events for a given 7-day window, that query would only need to access 7 out of 730 partitions, reducing the amount of data accessed by over 100x.
While static partitioning is very useful, there are several challenges with it. For example, a table with too many partitions can result in a significant amount of metadata to manage and maintain. Partitioning at too fine of a granularity can also limit query parallelism because the amount of data becomes too small in any given partition. And finally, there are a finite number of useful partitioning schemes as it is impossible to partition on everything.
To address these challenges, Apache Impala 2.9 added a number of features and functionality to provide even faster query performance using more advanced data skipping techniques.
Leveraging Parquet Column Statistics and Dictionary Filtering
Each Apache Parquet file contains a footer where metadata can be stored including information like the minimum and maximum value for each column. Starting in v2.9, Impala populates the min_value and max_value fields for each column when writing Parquet files for all data types and leverages data skipping when those files are read. This approach significantly speeds up selective queries by further eliminating data beyond what static partitioning alone can do. For files written by Hive / Spark, Impala only reads the deprecated min and max fields.
The effectiveness of the Parquet min_value/max_value column statistics for data skipping can be increased by ordering (or clustering1) data when it is written by reducing the range of values that fall between the minimum and maximum value for any given file. It was for this reason that Impala 2.9 added the SORT BY clause to table DDL which directs Impala to sort data locally during an INSERT before writing the data to files.
In addition to leveraging Parquet column statistics for data elimination, Impala also added optimizations for dictionary filtering. Low-cardinality columns are generally dictionary encoded so even if the min/max filtering does not disqualify the row group, dictionary predicate evaluation may often do so.
- Clustering here refers to the data organization, but Impala also has a
CLUSTEREDhint, see IMPALA-2521 ↩︎
Simple Illustrative Examples
Let’s walk through some simple illustrative examples to understand both Parquet row group skipping and dictionary filtering in action. These examples look at each independently, though it is possible for both to be used for a given query.
Example 1: Parquet Row Group Skipping via Min/Max Statistics
Say we have:
- A table that contains a string column named
letterwhich contains a single uppercase letter (‘A’ through ‘Z’) - Five Parquet files of data, each containing roughly the same number of rows
- All letters are present and equally represented in the data
- The query
select ... from t1 where letter = 'Q'
Now consider the following worst-case scenario where each of the five files contains both an ‘A’ and ‘Z’ value in the letter column.
| File# | min_value | max_value | Is ‘Q’ between min_value and max_value? |
|---|---|---|---|
| 1 | A | Z | YES |
| 2 | A | Z | YES |
| 3 | A | Z | YES |
| 4 | A | Z | YES |
| 5 | A | Z | YES |
When evaluating the letter = 'Q' predicate, the table scan will read the Parquet footer metadata, check the column statistics for letter and evaluate: Does ‘Q’ fall between min_value (‘A’) and max_value (‘Z’) for each file? For all five files in this case, ‘Q’ falls between ‘A’ and ‘Z’ so all data must then be read from each file.
Now let’s consider the best case scenario where the table was created with a SORT BY (letter) clause resulting in ordering by the letter column when data was loaded. In that case, the data layout may look like such:
| File# | min_value | max_value | Is ‘Q’ between min_value and max_value? |
|---|---|---|---|
| 1 | A | D | NO |
| 2 | E | J | NO |
| 3 | K | N | NO |
| 4 | O | S | YES |
| 5 | T | Z | NO |
With this data layout, the table scan now can evaluate the letter = 'Q' predicate and determine simply from reading the Parquet min/max column statistics in the footer if any rows in that file would match or not. For this example, we can see that four out of five files can now be skipped using this technique.
Example 2: Parquet Row Group Skipping via Dictionary Filtering
Much like Parquet row group skipping based on column min/max values, dictionary filtering can eliminate entire row groups based on single-column predicates. This works on any purely dictionary-encoded column. Impala uses dictionary encoding automatically, unless a column exceeds 40,000 distinct values in a given file. This means that when the min/max skipping may not be effective due to the min/max range being too large, dictionary filtering can check for the existence of values without reading any of the rows in the file.
For this example, we use data that contains a column containing two character state abbreviations. The table is populated via five insert statements thus creating five Parquet files. Four inserts include all states but CA, and the fifth includes all states including CA. This means that rows containing a value of CA only exist in one file. A query with the predicate ca_state = 'CA' can then leverage dictionary filtering to check for the existence of CA in the dictionary and completely skip reading the rows in four out of the five files.
Relevant Query Profile Information
The query execution plan now has annotations related to predicates that can be used for min/max and dictionary data skipping for Parquet files. For example, in the SCAN node we can now see the presence of both Parquet statistics and dictionary predicates (denoted by the <<< below). This is in addition to the table predicate.
00:SCAN HDFS [customer_address] partitions=1/1 files=5 size=42MB predicates: ca_state = 'CA' parquet statistics predicates: ca_state = 'CA' <<< parquet dictionary predicates: ca_state = 'CA' <<<
Additionally, new metrics have been added in the runtime profile to identify how many Parquet row groups get skipped using each of these techniques. These metrics are named NumDictFilteredRowGroups and NumStatsFilteredRowGroups and can be found in the SCAN node section for each impalad instance. To understand how effective the data skipping is for a given query, compare NumRowGroups (the total number of row groups) to the aforementioned statistics (the number of groups skipped).
For Example 2 above, the relevant statistics mentioned above are
Total NumRowGroups = 5
Total NumStatsFilteredRowGroups = 0
Total NumDictFilteredRowGroups = 4Partitioning vs. Clustering
To demonstrate the effectiveness of the SORT BY clause combined with Parquet data skipping, we use the TPC-DS table store_sales from the 1TB scale factor and create two versions, one that uses the PARTITIONED BY (ss_sold_date_sk) clause and one that uses the SORT BY (ss_sold_date_sk) clause. Both tables were then loaded via Impala from the same text file backed table and contain 2,879,987,999 rows.
The partitioned version of store_sales contains 1824 partitions, one for each unique value of ss_sold_date_sk. Each partition contains exactly one Parquet file ranging in size from 57MB to 265MB.
The clustered version of the table contains 574 files ranging in size from 10MB to 253MB. To be more precise, all but 14 of the files are 253MB.
Consider the following query:
select
avg(ss_wholesale_cost)
from
store_sales
where
ss_sold_date_sk = 2452590;It’s obvious that with partitioning, exactly one partition and thus one file will be accessed — so the bar is quite high, since only 1 file of 1824 would be accessed.
For the clustered table let’s start by analyzing the query profile. The relevant metrics are pulled out using a script (see below). We can see the following:
Total NumRowGroups = 574
Total NumStatsFilteredRowGroups = 535
Total NumDictFilteredRowGroups = 12As mentioned above, the clustered table consists of 574 files and given Impala writes one row group per file, there are 574 total row groups. Of those 574 files, 535 were able to be eliminated using the min/max filtering. Of the remaining 39 files/row groups, 12 more were able to be eliminated using dictionary filtering leaving just 27 files left to be scanned. Thus in total, 547 of 574 files (or 95.3%) were eliminated and no rows were scanned nor materialized from those files. This puts the performance of the query on the clustered table on par with that of the partitioned table since the files are read in parallel.
Examples of Combining Partitioning and Clustering
When partitioning and clustering are combined it can have a significant performance impact on queries. Below are two different use cases of combining the two features.
Example 1: Partitioning + Clustering on a Related Dimension
SQL queries over time-based data generally filter over a specific time range or window. Most time-based tables are partitioned by calendar date, or even more granular, but in this example, we demonstrate the impacts of using partitioning for coarse-grained elimination to the day and then adding in clustering to further organize the data by hour.
Below is the DDL for each of the three tables:
-- base table
create table store_sales_denorm_base (...);
-- partitioned table
create table store_sales_denorm_partition (...)
partitioned by (d_year int, d_moy int);
-- partitioned and clustered table
create table store_sales_denorm_partition_sorted (...)
partitioned by (d_year int, d_moy int)
sort by (d_date, d_dom, t_hour);Business question: Find top 10 customers by revenue who made purchases on Christmas Eve between 1am and noon.
SQL query:
select
sum(ss_ext_sales_price) as revenue,
count(ss_quantity) as quantity_count,
customer_name
from store_sales_x
where d_year = 2002
and d_moy = 12
and d_dom = 24
and t_hour between 1 and 12
group by customer_name
order by revenue desc
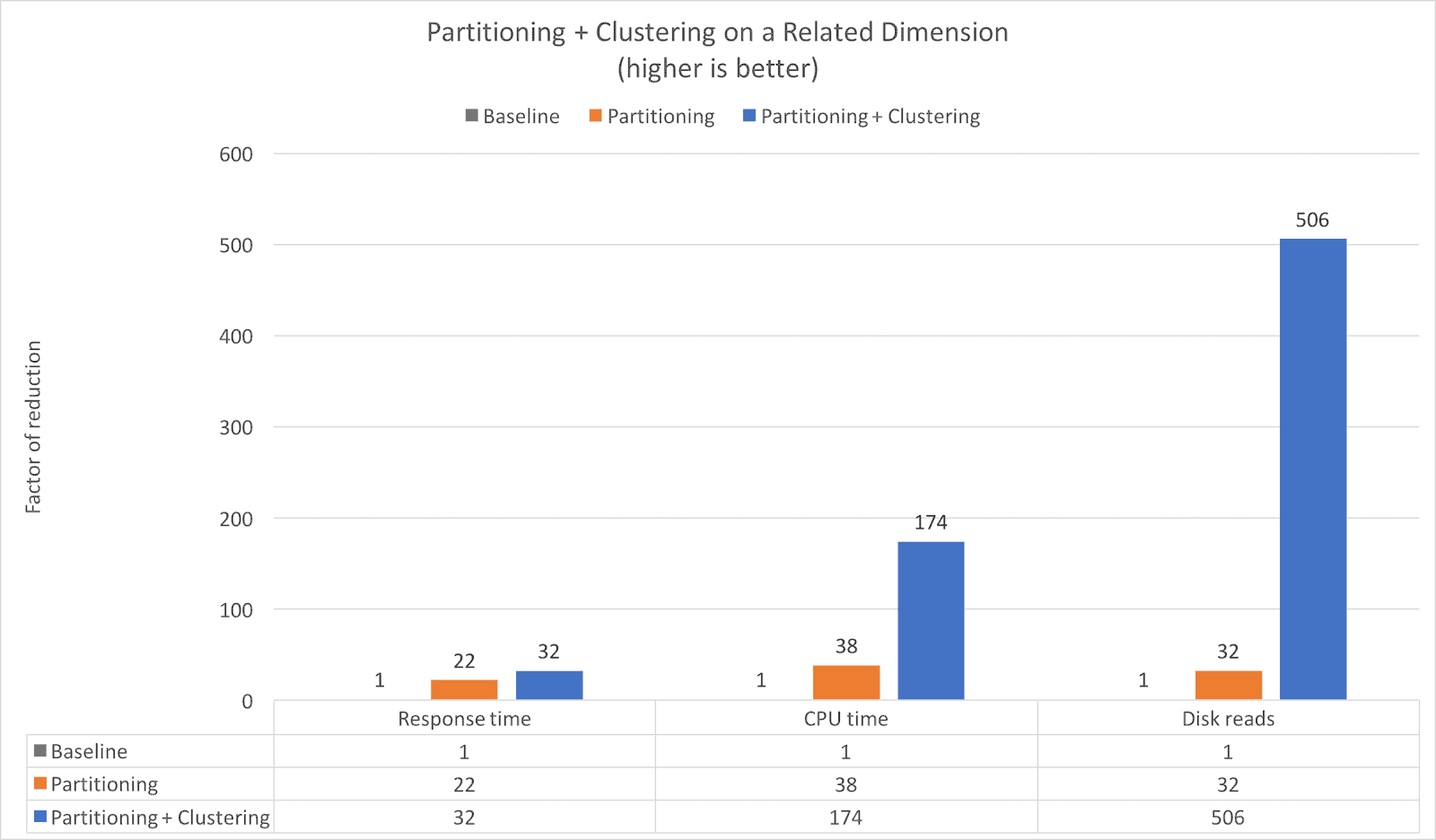
limit 10;As you can see from the chart, the response time is reduced by a factor of 22 by using day based partitioning, but when combined with clustering the response time is reduced by 32x compared to the base table. Additionally, you can see that both the CPU time and Disk reads are reduced significantly, even when comparing to the partitioned table. The data clustering reduces the CPU time by an additional 4.5x (38x to 174x) and the Disk reads by an additional 15.8x (32x to 506x).
Example 2: Partitioning + Clustering on a Non-related Dimension
In this example we once again use date based partitioning, but instead of using clustering on a related date/time dimension, a different and selective dimension/attribute is used for the clustering column. This approach greatly benefits use cases where there is a selective predicate, but there are too many distinct values to partition on that column effectively.
Below is the DDL for each of the three tables:
-- base table
create table store_sales_denorm_base (...)
-- partitioned table
create table store_sales_denorm_partition (...)
partitioned by (d_year int, d_moy int);
-- partitioned and clustered table
create table store_sales_denorm_partition_customer_sorted
partitioned by (d_year int, d_moy int)
sort by (customer_name, d_date, t_time, t_hour, t_minute);Business question: Find all the records from 2002 for the customer Meridith Smith.
SQL query:
select *
from store_sales_x
where customer_name = 'Meridith Smith'
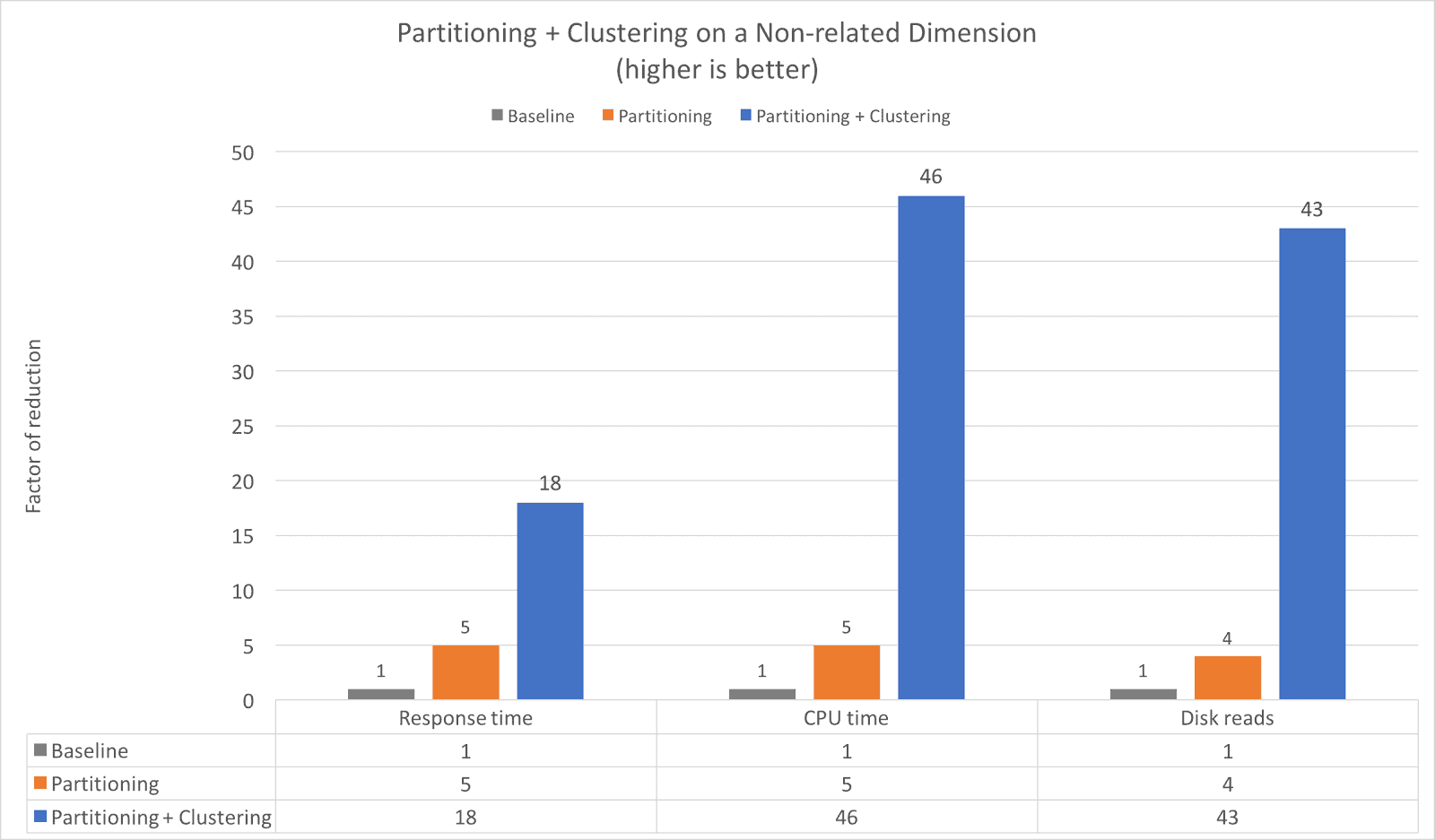
and d_year = 2002;Again, we see the impacts of clustering are quite significant compared to just using partitioning. The addition of clustering to the partitioned table further speeds up the query an additional 3.6x (from 5x to 18x) while reducing the CPU time 9.2x (from 5x to 46x) and reducing Disk reads over 10x (from 4x to 43x).
Future Work
- As of Impala 2.9, Runtime Filters do not yet support statistics predicates but this is something being worked on. See IMPALA-3430.
- Very selective queries often see a significant amount of time spent in tuple materialization. With the combination of min/max and dictionary skipping techniques, this should be much less of an issue since skipped row groups are never materialized. See IMPALA-2017.
Summary
Impala’s new data elimination technique works by applying predicates against Parquet column statistics and dictionaries. It complements the existing partitioning mechanism and further improves the performance of selective queries. Impala now populates column statistics during INSERT and provides a new SORT BY() clause during table creation to maximize the filtering effect on the selected columns.
Related JIRAs
For those interested in understanding more, feel free to research the following tickets and related code reviews:
- IMPALA-4624: Add dictionary filtering to Parquet scanner
- IMPALA-4166: Add SORT BY clause in CREATE TABLE statement
- IMPALA-4989: Improve filtering based on parquet::Statistics
- IMPALA-3430: Runtime filter : Extend runtime filter to support Min/Max values
- IMPALA-2017: Lazy materialization of columns during query
Bash Script
#!/bin/bash
# url to query profile
# notice the &raw at the end to return it in text format
URL="http://impalad:25000/query_profile?query_id=da452092e9b4181e:98bc374500000000&raw"
function getmetric {
# Grab the profile from the URL, find the relevant metric, and discard
# the first line since it is the average number across all instances.
# Add up the values (column 3) from rest.
echo -n "Total $1 = "
curl -sS $URL | grep $1 | sed 1d | awk '{sum += $3} END {print sum}'
}
for metric in NumRowGroups NumStatsFilteredRowGroups NumDictFilteredRowGroups
do
getmetric $metric
done