Small files are a common challenge in the Apache Hadoop world and when not handled with care, they can lead to a number of complications. The Apache Hadoop Distributed File System (HDFS) was developed to store and process large data sets over the range of terabytes and petabytes. However, HDFS stores small files inefficiently, leading to inefficient Namenode memory utilization and RPC calls, block scanning throughput degradation, and reduced application layer performance. In this blog post, we will define the issue of small file storage and examine ways to tackle it while keeping the complications at bay.
What are Small Files?
A small file is one which is significantly smaller than the default Apache Hadoop HDFS default block size (128MB by default in CDH). One should note that it is expected and inevitable to have some small files on HDFS. These are files like library jars, XML configuration files, temporary staging files, and so on. But when small files become a significant part of datasets, the problems arise. Hence, in this section, we shall discuss why it is a good goal to have a file size as close to a multiple of the HDFS block size as possible.
Hadoop’s storage and application layers are not designed to function efficiently with a large number of small files. Before we get to the implications of this, let’s review how HDFS stores files.
In HDFS, data and metadata are separate entities. Files are split into blocks that are stored and replicated on the DataNodes’ local file systems across the cluster. The HDFS namespace tree and associated metadata are maintained as objects in the NameNode’s memory (and backed up to disk), each of which occupies approximately 150 bytes, as a rule of thumb. This arrangement is described in more detail in the public documentation here.
The two scenarios below illustrate the small files issue:
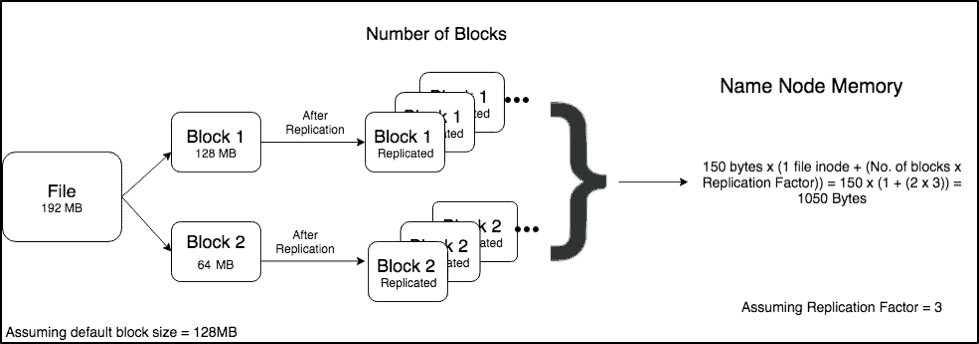
Scenario 1 (1 large file of 192MiB):
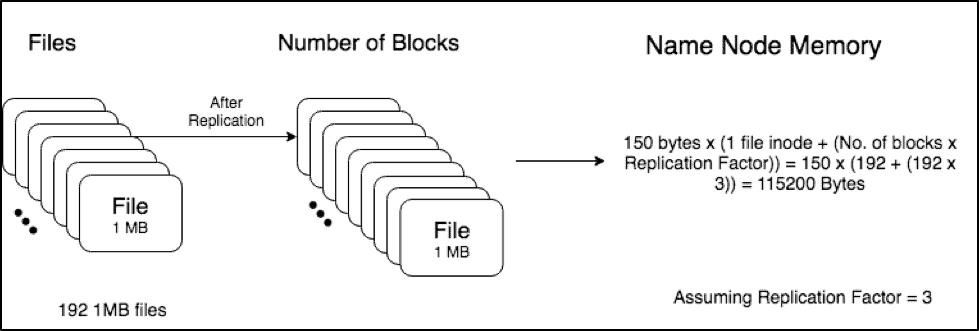
Scenario 2 (192 small files, 1MiB each):
Scenario 1 has one file which is 192MB which is broken down to 2 blocks of size 128MB and 64MB. After replication, the total memory required to store the metadata of a file is = 150 bytes x (1 file inode + (No. of blocks x Replication Factor)).
According to this calculation, the total memory required to store the metadata of this file on the Namenode = 150 x (1 + (2 x 3)) = 1050 Bytes.
In contrast, scenario 2 has 192 1 MB files. These files are then replicated across the cluster. The total memory required by the Namenode to store the metadata of these files = 150 x (192 + (192 x 3)) = 115200 Bytes.
Hence, we can see that we require more than 100x memory on the Namenode heap to store the multiple small files as opposed to one big 192MB file.
Effects on the Storage Layer
When a NameNode restarts, it must load the filesystem metadata from local disk into memory. This means that if the namenode metadata is large, restarts will be slower. The NameNode must also track changes in the block locations on the cluster. Too many small files can also cause the NameNode to run out of metadata space in memory before the DataNodes run out of data space on disk. The datanodes also report block changes to the NameNode over the network; more blocks means more changes to report over the network.
More files mean more read requests that need to be served by the NameNode, which may end up clogging NameNode’s capacity to do so. This will increase the RPC queue and processing latency, which will then lead to degraded performance and responsiveness. An overall RPC workload of close to 40K~50K RPCs/s is considered high.
Effects on Application Layer
In general, having a large number of small files results in more disk seeks while running computations through an analytical SQL engine like Impala or an application framework like MapReduce or Spark.
MapReduce/Spark
In Hadoop, a block is the most granular unit of data on which computation can be performed. Thus, it affects the throughput of an application. In MapReduce, an individual Map task is spawned for each block that must be read. Hence, a block with very little data can degrade performance, increase Application Master bookkeeping, task scheduling, and task creation overhead since each task requires its own JVM process.
This concept is similar for Spark, in which each “map” equivalent task within an executor reads and processes one partition at a time. Each partition is one HDFS block by default. Hence, a single concurrent task can run for every partition in a Spark RDD. This means that if you have a lot of small files, each file is read in a different partition and this will cause a substantial task scheduling overhead compounded by lower throughput per CPU core.
MapReduce jobs also create 0 byte files such as _SUCCESS and _FAILURE. These files do not account for any HDFS blocks but they still register as an inode entry in the Namenode heap which uses 150 bytes each as described earlier. An easy and effective way to clear these files is by using the below HDFS command:
hdfs dfs -ls -R | awk '$1 !~ /^d/ && $5 == "0" { print $8 }' | xargs -n100 hdfs dfs –rm
This will move those files to the .Trash location from where it will be cleared out automatically once the trash retention policy takes effect.
Note: This should not be done while your workloads are running on the specified path since it may cause applications to fail if they have dependencies on these files to know when the jobs complete or fail.
Impala—Effects on the Catalog Server
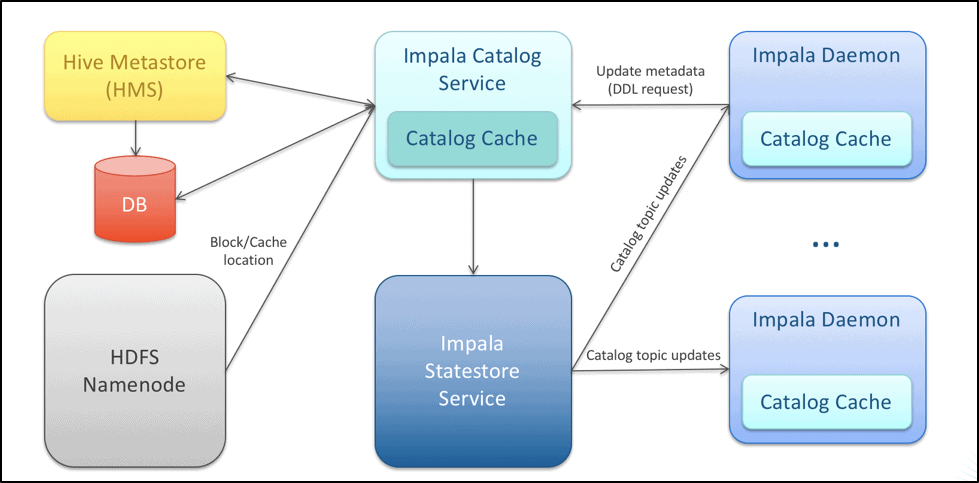
Impala is a high-performance query engine, which caches the HDFS namespace information in the Catalog Server for faster metadata access. Below is an architecture diagram detailing the way the Impala catalog is maintained and distributed across the service.
As seen with complications around NameNode metadata management, a similar issue arises with the metadata that Impala needs to maintain in the Catalog Server. The catalog size is a function of the number and size of objects maintained in the Catalog Server. These objects with their estimated average memory usage are described in the table below:
| Object | Memory Usage |
| Table | 5KB |
| Partition | 2KB |
| Column | 100B |
| Incremental Stats | 400B* (per column per partition) |
| File | 750B |
| File Block | 300B |
*Can go as high as 1.4KB/Column/Partition
Example: If there are 1000 tables with 200 partitions each and 10 files per partitions, the Impala Catalog Size will be at least (excluding table stats and table width):
#tables * 5KB + #partitions * 2kb + #files * 750B + #file_blocks * 300B = 5MB + 400MB + 1.5GB + 600MB = ~ 2.5GB
The larger the Impala Catalog Size the higher its memory footprint. Large metadata in the HMS for Hive/Impala is not advised as it needs to keep track of more files, causing:
- Longer Metadata loading time
- Longer StateStore topic update time
- Slow DDL statement operations
- Longer query plan distribution time
In addition to the issues related to the metadata, each disk read is single threaded by default in Impala which can cause a significant overhead in I/O with small files. Further, if the table is stored in the parquet file format, each physical file needs to be opened/closed twice; that is, once for the read footer and again for the column data.
How Do Small Files Originate?
Let us discuss some of the common mistakes that may give birth to insidious small files.
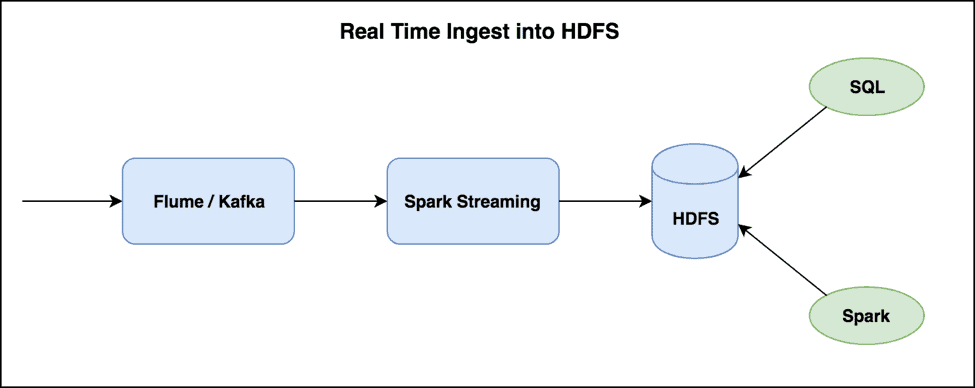
Streaming Data
Data ingested incrementally and in small batches can end up creating a large number of small files over a period of time. Near-real-time requirements for streaming data, with small windows (every few minutes or hours) that do not create much data will cause this problem. Below is a typical streaming ETL ingest pipeline into HDFS.
Large Number of Mappers/Reducers
MapReduce jobs and Hive queries with large number of mappers or reducers can generate a number of files on HDFS proportional to the number of mappers (for Map-Only jobs) or reducers (for MapReduce jobs). Large number of reducers with not enough data being written to HDFS will dilute the result set to files that are small, because each reducer writes one file. Along the same lines, data skew can have a similar effect in which most of the data is routed to one or a few reducers, leaving the other reducers with little data to write, resulting in small files.
Over-Partitioned Tables
An over-partitioned table is a partitioned Hive table with a small amount of data (< 256 MB) per partition. The Hive Metastore Server (HMS) API call overhead increases with the number of partitions that a table maintains. This in return leads to deteriorated performance. In these cases, consider reviewing the partition design and reducing the partition granularity, for example from daily to monthly partitions.
Over-Parallelizing
In a Spark job, depending on the number of partitions mentioned in a write task, a new file gets written per partition. This is similar to having a new file getting created for each reduce task in the MapReduce framework. The more Spark partitions, the more files are written. Control the number of partitions to curb the generation of small files.
File Formats and Compression
Using of inefficient file formats, for example TextFile format and storing data without compression compounds the small file issue, affecting performance and scalability in different ways:
- Reading data from very wide tables (tables with a large number of columns) stored as non-columnar formats (TextFile, SequenceFile, Avro) requires that each record be completely read from disk, even if only a few columns are required. Columnar formats, like Parquet, allow the reading of only the required columns from disk, which can significantly improve performance
- Use of inefficient file formats, especially uncompressed ones, increases the HDFS space usage and the number of blocks that need to be tracked by the NameNode. If the files are small in size, it means the data is split into a larger number of files thereby increasing the amount of associated metadata to be stored.
Identifying Small Files
FSImage and fsck
Because the NameNode stores all the metadata related to the files, it keeps the entire namespace image in RAM. This is the persistent record of the image stored in the NameNode’s local native filesystem – fsimage. Thus we can analyze the fsimage or the fsck output to identify paths with small files.
The fields available in the fsimage are:
Path, Replication, ModificationTime, AccessTime, PreferredBlockSize, BlocksCount, FileSize, NSQUOTA, DSQUOTA, Permission, UserName, GroupName
The fsimage can be processed in an application framework like MapReduce or Spark and even loaded into a Hive table for easy SQL access.
Another approach is using the fsck output and parsing that to load it into a Hive table for analysis. There are a few variants of this approach; here is a public project that uses PySpark and Hive to achieve this. It aggregates the total number of blocks, average block size and total file size at each HDFS path which can then be queried in Hive or Impala.
Cloudera Navigator
Cloudera Navigator is a data governance product with audit, lineage, metadata management, data stewardship and policy enforcement features.
The Navigator Search and Analytics tabs can be used to identify small files easily. The HDFS search filters in the left panel allows to filter for files under a specific size or range. The new version of Cloudera Navigator (2.14.x) even has an in-built Dashboard widget to identify small files as shown below.
Ways to Tackle Small Files
Preventative
Streaming Ingest Use-Case
As mentioned earlier, ingesting streaming data usually leads to creating small files. Tweaking the rate of ingest, window, or dstream size (Spark) can help alleviate some of the issues. But usually to meet near-real-time analytics demands, some architectural changes need to be introduced in the HDFS ingestion pipeline with respect to intermediate compaction jobs, maintaining multiple landing directories, and active/passive versions of table data. This is discussed in more detail in this Cloudera Engineering blog.
For near-real-time analytical processing, HBase and Kudu are better choices for storage layers, based on the data type (unstructured vs structured), append/update frequency and data usage patterns (random reads vs aggregations).
Batch Ingest Use-Case
For batch ingest pipelines, a good choice is a regularly scheduled compaction job, which compacts files after landing into HDFS. The file compaction tools mentioned later in this blog would be good candidates for this.
Over-Partitioned Tables
We should aim to have partitions with a significant volume of data so that the files within each partition are large. While deciding on the granularity of the partitions, consider the volume of data that will be stored per partition. Plan for partitions that have large files (~256MB or larger with Parquet), even if it means having less granular partitions, such as monthly instead of daily. For example, keeping the number of partitions within 10K-30K during the lifetime of a table is a good guideline to follow.
For tables that have small data volumes (few hundred MBs), consider creating a non-partitioned table. It can be more efficient to scan all the (small) table’s data stored in a single file than having to deal with thousands of files scattered throughout multiple partitions with tiny number of bytes.
Creating buckets for your table can also reduce the number of small files by essentially fixing the number of reducers and output files generated.
Spark Over-Parallelizing
When writing data to HDFS in Spark, repartition or coalesce the partitions before writing to disk. The number of partitions defined in those statements will determine the number of output files. Checking the output of the Spark Job and verifying the number of files created and throughput achieved is highly recommended.
Prescriptive
HDFS File Compaction Tools
The most obvious solution to small files is to run a file compaction job that rewrites the files into larger files in HDFS. A popular tool for this is FileCrush. There are also other public projects available such as the Spark compaction tool.
Re-Create Table in Hive
To ensure a good balance between performance and efficient storage, create tables using the PARQUET file format and ensure that data compression is enabled when writing data to them.
If you have an existing Hive table that has a large number of small files, you can re-write the table with the below configuration settings applied before re-writing:
set hive.exec.compress.output=true; set hive.exec.parallel = true; set parquet.compression=snappy; set hive.merge.mapfiles=true; set hive.merge.mapredfiles=true; set hive.merge.smallfiles.avgsize = 134217728; --128M set hive.merge.size.per.task = 268435456; --256M set hive.optimize.sort.dynamic.partition = true; set parquet.blocksize= 268435456; --256M set dfs.block.size=268435456; --256M
Note: The average size and parquet block sizes specified here are for representation purposes only and should be changed based on the application and needs. Details on the Hive configuration properties can be found on the official Apache Hive page.
There are two ways to do this:
- You can run a CREATE TABLE AS SELECT (CTAS) statement to create the target table, as long as the target table is not partitioned, is not external, and is not bucketed.
- To overcome those limitations, instead of a direct CTAS, you can run a CREATE TABLE LIKE (CTL) statement to copy the source table schema to create the target table and then use an INSERT OVERWRITE SELECT statement to load the data from the source table to the target table.
Note: you will need to enable non-strict dynamic partition mode in Hive if the data is being inserted without a static partition name defined. This can be done by settinghive.exec.dynamic.partition.mode=nonstrict
The partition column(s) must be the last column(s) in the select statement for dynamic partitions to work in this context.
Consider the following simplified example:
create external table target_tbl like source_tbl stored as parquet location '; set hive.exec.dynamic.partition.mode=nonstrict; insert overwrite table target_tbl partition (partition_col) select * from source_tbl;
Similar CTAS can be executed in Impala as well, but if the query runs with multiple fragments on different nodes you will get one file per fragment. To avoid this, you could restrict Impala to run the query on a single node using set num_nodes=1 but this approach is not recommended since it removes parallelism and causes slow inserts, degrading the performance, and could cause the daemon to run out of memory if writing a large table.
Additionally, the number of reducers can be configured directly as well using the mapred.reduce.tasks setting. The number of files created will be equal to the number of reducers used. Setting an optimal reducer value depends on the volume of the data being written.
Conclusion
Prevention is better than cure. Hence, it is critical to review application design and catch users in the act of creating small files. Having a reasonable number of small files might be acceptable, but too many of them can be detrimental to your cluster. Eventually leading to irritation, tears, and extended hours at work. Therefore, Happy Cluster, Happy Life!
Have any questions or want to connect with other users? Check out the Cloudera Community
Shashank Naik is a Senior Solutions Consultant at Cloudera.
Bhagya Gummalla is a Solutions Consultant at Cloudera.




Nordos technology encapsulates big number of small files and easy gives you out-of the box solution as a on prem solution that you can use as a back-end for hdfs/hadoop. It is developed in C for linux and windows. It allows to estimate total size that will be used in File System, reconstruct clsuters on the fly, has variable adaptable data block that is used according incoming data
So when doing the re-create (CTL) you are are going to a new table requiring a switch. Some examples write to same table – if so, would there bean issue with concurrency?