Thanks to Karthik Vadla, Abhi Basu, and Monica Martinez-Canales of Intel Corp. for the following guest post about using CDH for cost-effective processing/indexing of DICOM (medical) images.
Medical imaging has rapidly become the best non-invasive method to evaluate a patient and determine whether a medical condition exists. Imaging is used to assist in the diagnosis of a condition and, in most cases, is the first step of the journey through the modern medical system. Advances in imaging technology has enabled us to gather more detailed, higher-resolution 2D, 3D, 4D, and microscopic images that are enabling faster diagnosis and treatment of certain complex conditions.
A high-resolution microscopic scan of a human brain can be as large as 66TB. Examples of some 3D modalities like computer tomography (CT) is about 4MB per image (2048×2048 pixels). We can appreciate medical imaging becoming a Big Data problem based on the fact that there are approximately 750PB of medical images in all the repositories in the United States, with that volume projected to cross 1EB (exabyte) by end of 2016.
Currently, Picture Archiving and Communication Systems (PACS), which provides proprietary data formats and objects, are the industry standard for storage and retrieval of medical imaging files. In this post, we describe an alternate solution for storing and retrieving medical imaging files on an Apache Hadoop (CDH) cluster to provide high-performance and cost-effective distributed image processing. The advantages of such a solution include the following:
- Allows organizations to bring their medical imaging data closer to other sources already in use like genomics, electronic medical records, pharmaceuticals, wearable data, and so on, reducing cost of data storage and movement
- Enables solutions with lower cost using open source software and industry-standard servers
- Enables medical image analytics and correlation of many forms of data within the same cluster
In this post, we will help you replicate Intel’s work as a proof point to index DICOM images for storage, management, and retrieval on a CDH cluster, using Apache Hadoop and Apache Solr.
Solution Overview
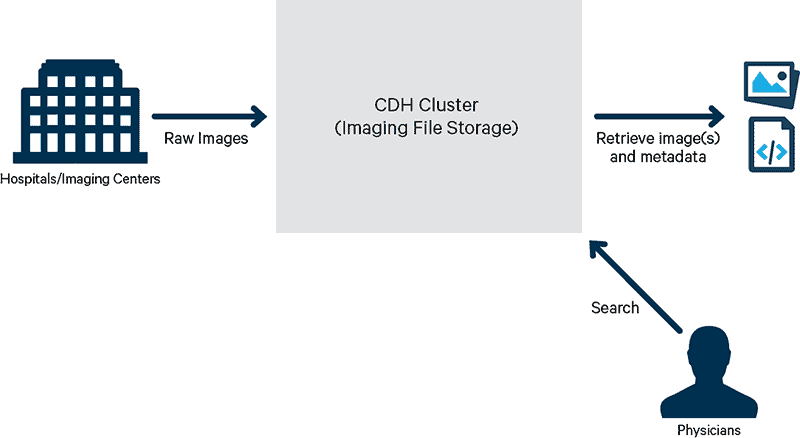
Let’s focus on a simple but essential example use case: the ability to store and retrieve DICOM images. This use case assumes no dependency on other existing image repositories. Hospitals and imaging facilities send copies of DICOM images to a HIPAA-compliant cloud or locally hosted server cluster where the images would be stored. On demand, the physician or caregiver should be able to search and retrieve patient DICOM images from the remote cluster connected to a local machine for viewing, as shown in Figure 1.
Figure 1
Details
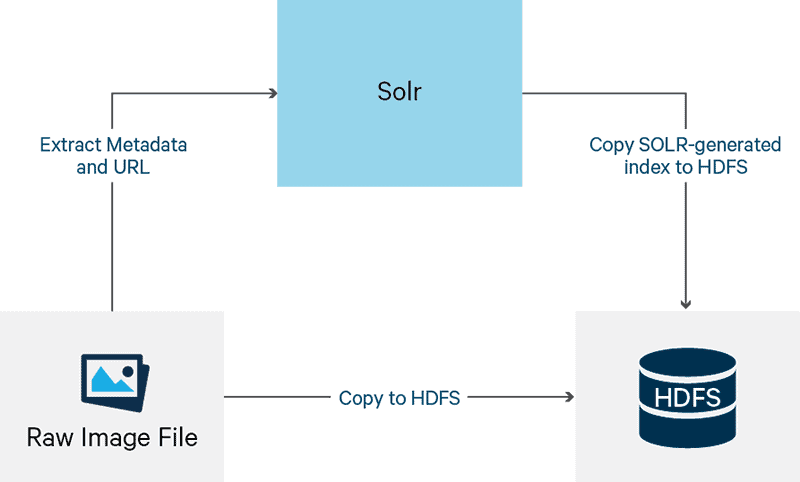
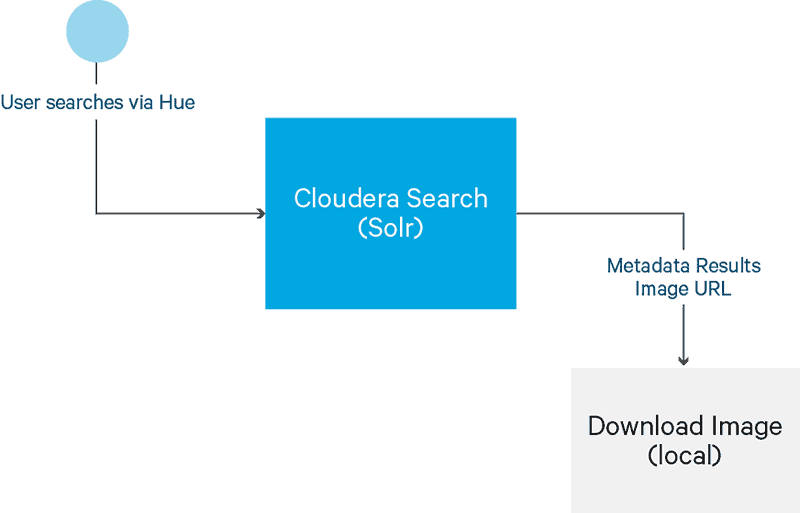
DICOM images consist of two sections: text header and binary image. To ensure images are stored and searchable as shown in Figure 2 and Figure 3, use the steps outlined below.
Figure 2
Figure 3
Note that Figure 2 represents the perspective of a software developer, while Figure 3 represents the perspective of an end user interacting with the architected solution. As software developers, we must go through this process ourselves to validate functionality for the end user.
- Extract metadata from the DICOM image.
- Store the original DICOM image on HDFS.
- Use metadata to generate an index file (that will also reside on the HDFS).
- Search using the Hue interface to retrieve an image.
- Download the image to the local system and open/view it using a DICOM viewer.
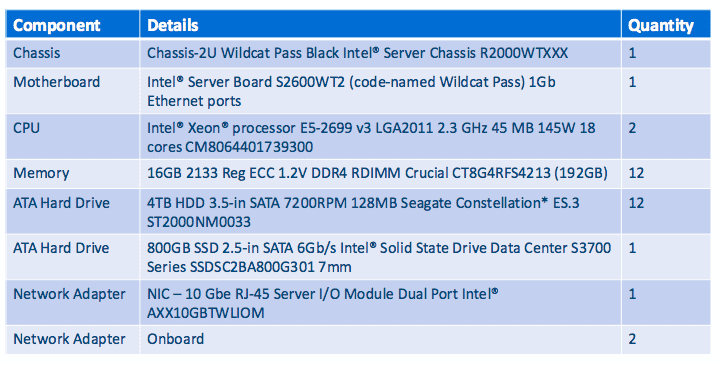
Industry-standard servers are flexible in terms of performance and storage when compared to the cost per node of high-performance server nodes. Our server nodes each used a 2-CPU Intel® Xeon® processor E5 family, 4TB x 12 (48TB) storage, and 192GB RAM. Processing and storage needs can be “horizontally scaled” by increasing the number of nodes in the cluster when required.
System Setup and Configuration
Listed below are the key specs of our test six-node CDH cluster and one edge node.
Software Requirements
To test this solution, we used CDH 5.4 with the following services enabled in Cloudera Manager:
- HDFS
- To store input DICOM XML files that will be indexed
- To store output indexed results
- Apache Solr (Solrcloud mode)
- Uses
schema.xmlto index given input DICOM XML files
- Uses
- Apache ZooKeeper
- SolrCloud mode uses ZooKeeper for distributed indexing
- Hue (Search feature enabled)
- Graphical interface to view Solr indexed results, where you can search DICOM images based on indexed fields
- Cloudera Search
- Using
MapReduceIndexerToolfor offline batch indexing
- Using
Dataset
Our test data set consisted of DICOM CT images that were downloaded from the Visible Human Project.
Workflow
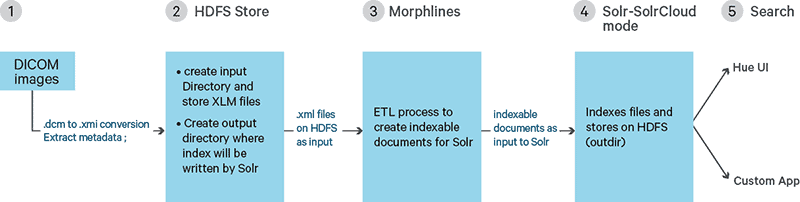
Figure 4 describes the workflow approach.
Figure 4
Step 1: Assuming you have stored all DICOM images in a local folder, extract DICOM metadata from images using the DCM Toolkit (DCMTK) with the dcm2xml utility and store them in XML format. (See Appendix 1 for an example of such a file.)
Example: dcm2xml ./dcm2xml source.dcm source.xml
In the above example, for dcm2xml utility to work, you have to set the path as below in .bashrc after downloading DCMTK to your local machine from where you want to run this script.
Example: export DCMDICTPATH="/home/root/Dicom_indexing/dicom-script/dcmtk-3.6.0/s
Step 2: Import the converted DICOM images to HDFS at /user/hadoop/input-dir and also create/user/hadoop/output-dir to store indexed results.
Step 3: Do ETL processing. For this step, a good approach is to use a Morphlines configuration file, which parses and extracts the required files as per our requirement and creates indexable documents for Solr.
Step 4 (see process below): Solr uses schema.xml configuration to determine which fields to index from a given XML file. We have also used MapReduceIndexerTool for this offline batch indexing. (See Appendix 2.)
- First, confirm that your Solr service (in SolrCloud mode) on the cluster is up and running (visit http://<your –solr-server-name>:8983/solr).
- Generate a configuration file using
solrctland include aschema.xmlwith the fields that you want to index (using the-- zkoption will provide ZooKeeper hostnames; you can find that info in Cloudera Manager’s ZooKeeper Service). The last entry of IP address needs the ZooKeeper port. Note: replace hostip with your ZooKeeper host IPs.solrctl instancedir --zk hostip1,hostip2,hostip3:2181/solr --generate $HOME/solr_config cd $HOME/solr_config/conf
Download the
schema.xmlfile to your local machine and edit it. Include all field names that you want to index. Field names should match the name attribute in your XML files to index. In the example here, the DICOM XML file contains hundreds of fields, of which we are going to index only about 10–15 fields. Update the modifiedschema.xmlto your/conffolder. (See Appendix 1 to see one of the generated DICOM XML file, and Appendix 2 to see the customizedschema.xml.) - Clean up any collections and instance directories existing in ZooKeeper:
solrctl --zk hostip1,hostip2,hostip3:2181/solr collection --delete demo-collection >& /dev/null solrctl --zk hostip1,hostip2,hostip3:2181/solr instancedir --delete demo-collection >& /dev/null
- Upload the Solr configuration to SolrCloud.
solrctl --zk hostip1,hostip2,hostip3:2181/solr instancedir --create demo-collection $HOME/solr_configs
- Create a Solr collection named
demo-collection.-s 2indicates that this collection has two shards.solrctl --zk hostip1,hostip2,hostip3:2181/solr collection --create demo-collection -s 2 -r 1 -m 2
- Create a directory to which
MapReduceBatchIndexercan write results. Verify it is empty.hadoop fs -rm -f -skipTrash -r output-dir hadoop fs -mkdir -p output-dir
- ETL processing—again, a good approach is to use a Morphlines configuration file. (See Appendix 3.)
- Use
MapReduceIndexerToolto index the data and push it live to demo-collection.${DICOM_WORKINGDIR}is the location wherelog4j.propertiesandmorphlines.confcan be found).${HDFS_DICOM_OUTDIR} - Location of output dir folder on hdfs (ex: /user/hadoop/output-dir) ${HDFS_DICOM_INDIR} – Location of input dir folder on hdfs (ex: /user/hadoop/input-dir/)hadoop \ jar \ ${Solr_HOME}/contrib/mr/search-mr-*-job.jar org.apache.solr.hadoop.MapReduceIndexerTool \ -D 'mapred.child.java.opts=-Xmx500m' \ --log4j ${DICOM_WORKINGDIR}/log4j.properties \ --morphline-file ${DICOM_WORKINGDIR}/morphlines.conf \ --output-dir ${HDFS_DICOM_OUTDIR} \ --verbose \ --go-live \ --zk-host hostip1,hostip2,hostip3:2181/solr \ --collection demo-collection \ ${HDFS_DICOM_INDIR}/dicomimagesSolr stores the indexed results in
output-dircreated above.
Step 5: Indexed results can be viewed using Hue; the DICOM image URL can be used to download image locally for viewing.
Testing
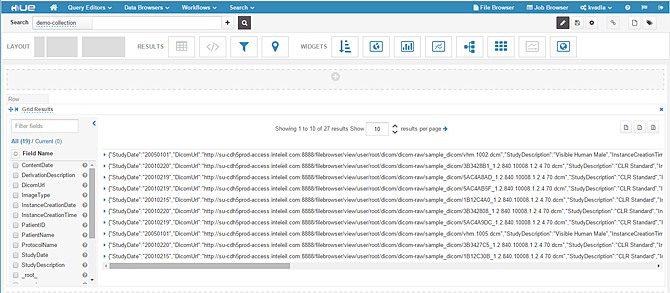
To test the indexed results, log in to your Hue interface. (We assume you already have access to Hue with the Search feature enabled in it.)
- Hover on Search and navigate to -> Indexes-> demo-collection->Search. Below is the default view of indexed results.
- Type a patient name or patient id or any field data that you have indexed. In this example, we are typing a patient name and other patient id, highlighted in the following screenshot.
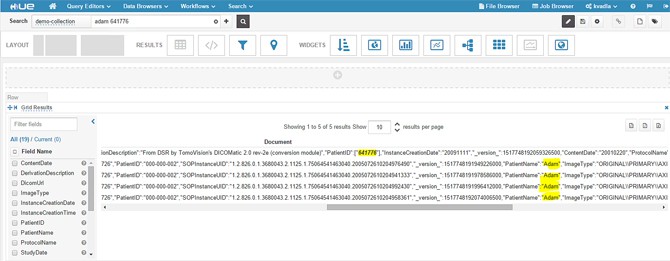
- When you expand a single result, you should see metadata fields as shown in the following screenshot.
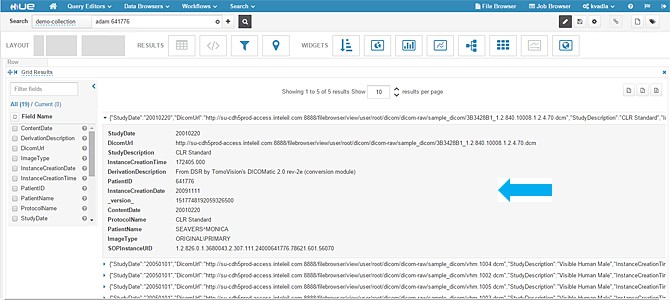
- In the above image you cannot click the DICOM URL. Instead, we used Hue to build a nice dashboard using some graphing widgets and added a clickable URL.
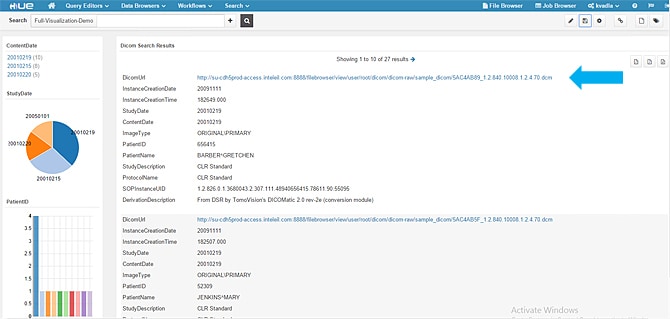
- When you click the DICOM URL, it gives you the option to download the
.dcmfile to your local machine. In this version, we are downloading to the local machine and viewing it with an open source tool called MicroDicom Viewer.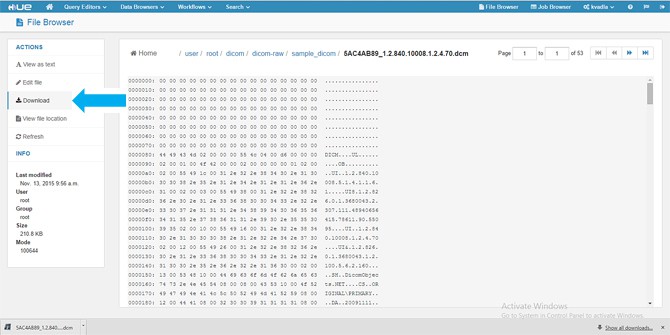
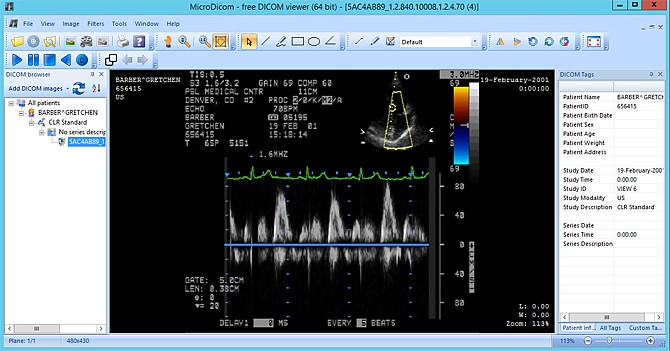
- View the image using the MicroDicom Viewer.
Future Work
We plan to continue to develop this reference architecture to allow a more streamlined method of downloading the DICOM file directly within the browser, using a plug-in. We will also be working toward better visualization capabilities and supporting multiple images to be downloaded simultaneously.
Karthik Vadla is a Software Engineer in the Big Data Solutions Pathfinding Group at Intel Corporation.
Abhi Basu is a Software Architect in the Big Data Solutions Pathfinding Group at Intel Corporation.
Monica Martinez-Canales is a Principal Engineer in the Big Data Pathfinding Group at Intel Corporation.
Appendix 1
216 1.2.840.10008.5.1.4.1.1.6.1 1.2.826.0.1.3680043.2.307.111.48712655111.78571.301.34207 1.2.840.10008.1.2.4.70 1.2.826.0.1.3680043.1.2.100.5.6.2.160 DicomObjects.NET ORIGINAL\PRIMARY 20091111 164835.000 1.2.826.0.1.3680043.2.307.111 1.2.840.10008.5.1.4.1.1.6.1 1.2.826.0.1.3680043.2.307.111.48712655111.78571.301.34207 20010215 20010215 093006.000 US CLR Standard From DSR by TomoVision's DICOMatic 2.0 rev-2e (conversion module) BURRUS^NOLA 655111 CLR Standard ………………………………………
Appendix 2
Mention the unique key along with this
SOPInstanceUID (Remove any previously existing unique key tag and replace with this tag.)
Appendix 3
Update your Morphline as below:
SOLR_LOCATOR : {
#This is the name of the collection which we created with solrctl utility in our earlier steps
collection : demo-collection
#Zookeeper host names, you will find this information in Cloudera Manager at ZooKeeper service
zkHost : "hostip1:2181, hostip2:2181, hostip3:2181/solr"
}
And include this specific XQuery inside the commands tag of morphlines
xquery {
fragments : [
{
fragmentPath : "/"
queryString : """
for $data in /file-format/data-set
return
{$data/element[@name='SOPInstanceUID']}
{$data/element[@name='ImageType']}
{$data/element[@name='InstanceCreationDate']}
{$data/element[@name='InstanceCreationTime']}
{$data/element[@name='StudyDate']}
{$data/element[@name='ContentDate']}
{$data/element[@name='DerivationDescription']}
{$data/element[@name='ProtocolName']}
{$data/element[@name='PatientID']}
{$data/element[@name='PatientName']}
{$data/element[@name='StudyDescription']}
{$data/element[@name='DicomUrl']}
"""
}
]
}
}