Cloudera Data Science Workbench provides freedom for data scientists. It gives them the flexibility to work with their favorite libraries using isolated environments with a container for each project.
In JVM world such as Java or Scala, using your favorite packages on a Spark cluster is easy. Each application manages preferred packages using fat JARs, and it brings independent environments with the Spark cluster. Many data scientists prefer Python to Scala for data science, but it is not straightforward to use a Python library on a PySpark cluster without modification. To solve this problem, data scientists are typically required to use the Anaconda parcel or a shared NFS mount to distribute dependencies. In the world of Python, it is standard to install packages with virtualenv/venv to isolated package environments before running code on their computer . Without virtualenv/venv, packages are directly installed on system directory, using virtualenv/venv makes sure to manage search path of appropriate packages in the specific directory. Supporting virtualenv is discussed on this JIRA, but basically, virtualenv is not something Spark will manage.
In this post, I will explain how to distribute your favorite Python library on PySpark cluster on The Cloudera Data Science Workbench Note that this technique is not the standard way in Python world. The original idea is written in this article.
The Goal
The goal of this article is to run Python code which uses a pure Python library on a distributed PySpark cluster.
Example code:
import os
import sys
from pyspark.sql import SparkSession
spark = SparkSession.builder \
.appName("spark-nltk") \
.getOrCreate()
data = spark.sparkContext.textFile('1970-Nixon.txt')
def word_tokenize(x):
import nltk
return nltk.word_tokenize(x)
def pos_tag(x):
import nltk
return nltk.pos_tag([x])
words = data.flatMap(word_tokenize)
words.saveAsTextFile('nixon_tokens')
pos_word = words.map(pos_tag)
pos_word.saveAsTextFile('nixon_token_pos')
The whole code is in the GitHub repository.
https://github.com/chezou/NLTK-pyspark
This code uses NLTK, Python’s natural language processing library. NLTK is not installed with conda by default. By adding `import` within your Python UDFs, you can use Python libraries. So let’s distribute NLTK with conda environment.
In this approach, libraries which require C extension might not work. I recommend creating conda recipe for libraries based on C extension. I will write a future article.
Create and pack conda environment
You can pack the python environment into the conda environment with `conda create` command. Open your workbench and run the following on your CDSW terminal:
$ conda create -n nltk_env --copy -y -q python=2 nltk numpy pip
$ source activate nltk_env (nltk_env)$ pip install some-awesome-package (nltk_env)$ cp -r ~/.local/lib ~/.conda/envs/nltk_env/
If you want to add extra pip packages without conda, you should copy packages manually after using `pip install`. In Cloudera Data Science Workbench, pip will install the packages into `~/.local`.
Be careful with using the `–copy` option which enables you to copy whole dependent packages into a certain directory of the conda environment.
Then Zip the conda environment for shipping on PySpark cluster.
$ cd ~/.conda/envs $ zip -r ../../nltk_env.zip nltk_env
(Optional) Prepare additional resources for distribution
If your code requires additional local data sources, such as taggers, you can both put data into HDFS and distribute archiving those files. For the sample code, you can use NLTK tokenizers and taggers which are assumed to be set on a local disk specified with environmental variables.
$ cd ~/ $ source activate nltk_env # download nltk data (nltk_env)$ python -m nltk.downloader -d nltk_data all (nltk_env)$ hdfs dfs -put nltk_data/corpora/state_union/1970-Nixon.txt ./ # archive nltk data for distribution (nltk_env)$ cd ~/nltk_data/tokenizers/ (nltk_env)$ zip -r ../../tokenizers.zip * (nltk_env)$ cd ~/nltk_data/taggers/ (nltk_env)$ zip -r ../../taggers.zip *
Put spark-defaults.conf on project root
You can set spark-submit option in spark-defaults.conf. Here is an example:
spark.yarn.appMasterEnv.PYSPARK_PYTHON=./NLTK/nltk_env/bin/python spark.yarn.appMasterEnv.NLTK_DATA=./ spark.executorEnv.NLTK_DATA=./ spark.yarn.dist.archives=nltk_env.zip#NLTK,tokenizers.zip#tokenizers,taggers.zip#taggers
By setting `spark.yarn.dist.archives=nltk_env.zip#NLTK`, PySpark unzips nltk_env.zip into the `NLTK` directory. `NLTK_DATA` is the environmental variable where NLTK data is put.
Set the environment variable
You should set the environment variable `PYSPARK_PYTHON` as `./NLTK/nltk_env/bin/python`. It enables PySpark to use Python within the conda environment.
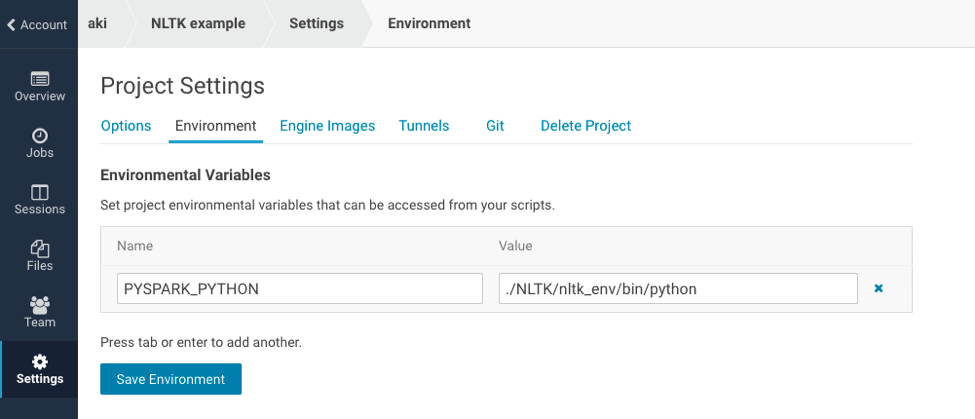
After setting environment variables, you should restart your session.
Run your script on CDSW
Now is the time to run your Python script. Open your workbench and run nltk_sample.py.
After running that, you can check the result as follows:
!hdfs dfs -cat ./nixon_tokens/* | head -n 10 Annual Message To The Congress On The State Of The ! hdfs dfs -cat nixon_token_pos/* | head -n 10 [(u'Annual', 'JJ')] [(u'Message', 'NN')] [(u'to', 'TO')] [(u'the', 'DT')] [(u'Congress', 'NNP')] [(u'on', 'IN')] [(u'the', 'DT')] [(u'State', 'NNP')] [(u'of', 'IN')]
Summary
Creating conda environment enables you to distribute your favorite Python packages without manual IT intervention using the Data Science Workbench tool by Cloudera. Data scientists can run their favorite packages without modifying your cluster.
To learn more about the Data Science Workbench visit our website.
Aki Ariga is a Field Data Scientist at Cloudera and sparklyr contributor




Hi Aki
Thanks for the instructions which work without problems in the built-in Workbench editor.
When following the same steps from a Jupyter notebook, I get an error on the executors, stating that the module was not distributed:
‘ModuleNotFoundError: No module named ‘nltk’
Do you have any idea what needs to be done additionally to do this from a Jupyter notebook? Thanks!
Best,
Matthias